ECP MEG Data Folders: Difference between revisions
Jump to navigation
Jump to search
| (9 intermediate revisions by the same user not shown) | |||
| Line 20: | Line 20: | ||
The data is labelled with Subjid+_+DataType+_run+RunNumber+_raw.fif | The data is labelled with Subjid+_+DataType+_run+RunNumber+_raw.fif | ||
[[file: RAW format.PNG | 300px]] | [[file: RAW format.PNG | 300px]] | ||
===Processing=== | ===Processing=== | ||
====Initialization==== | ====Initialization - Subject Dataframe ==== | ||
init_subject_dataframe - initialization creates a subject pandas dataframe saved as a csv file in the aProjects folder at the top of the MEG Project folder | init_subject_dataframe - initialization creates a subject pandas dataframe saved as a csv file in the aProjects folder at the top of the MEG Project folder | ||
[[file:AProject Folder.png|400px]] | [[file:AProject Folder.png|400px]] | ||
<br> | <br> | ||
The Contents of the subject dataframe are stored in a .csv file which can be opened using LibreOffice | The Contents of the subject dataframe are stored in a .csv file which can be opened using LibreOffice | ||
The data file is stored in the row, and all of the documented processing steps are the columns | |||
[[file:Subject DataFrame.PNG|1400px]] | [[file:Subject DataFrame.PNG|1400px]] | ||
==== | ====Initialization - Subject Processing Folders==== | ||
init_subject_process - | init_subject_process - A process folder is created for each dataset type (multiple runs are copied into each folder) | ||
====Cleaning==== | The _tsss.fif files are links to the tSSS folder. If these folders are deleted, it will not affect the data in the tSSS folder | ||
[[file:Process setup.png|300px]] | |||
=====Cleaning===== | |||
Cleaning (baddata pipeline) performs ICA removal of heartbeat and eyeblink artifact that is correlated to EOG and ECG electrodes at a threshold of (r=???) | Cleaning (baddata pipeline) performs ICA removal of heartbeat and eyeblink artifact that is correlated to EOG and ECG electrodes at a threshold of (r=???) | ||
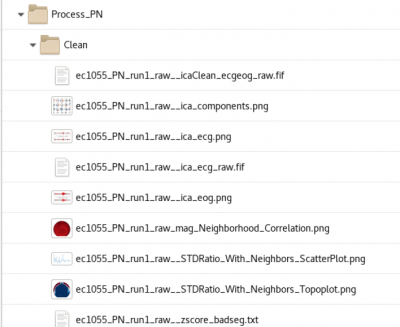
The processing creates a folder called Process_*DataType*/Clean with the output of the baddata analysis | |||
[[file:Clean data.png|400px]] | [[file:Clean data.png|400px]] | ||
Latest revision as of 20:19, 10 June 2019
At the MEG lab:
Data is acquired to the MEG_acq/ecp/ folder Data is tSSS processed and organized into raw and tSSS folders (the format of the data can be seen below)Data is uploaded to to the RCC using scp (/rcc/stor1/projects/ECP/MEG/MEG_Orig) Data is copied into /rcc/stor1/projects/ECP/MEG/MEG_Work for further processing
Folder Layout /rcc/stor1/projects/ECP/MEG/MEG_Work
All subjects have a folder with the following folders: Anatomy, RAW, tSSS, Process_*DataType*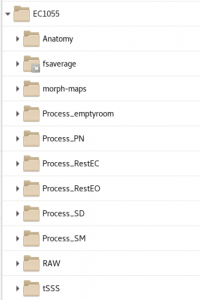
Anatomy Layout
Typical Freesurfer layout with BEM process performed The folder is always labelled Anatomy (a hack to make all folders homogeneous and easier to process) The Anatomy-trans.fif is created during the MNE python coregistration step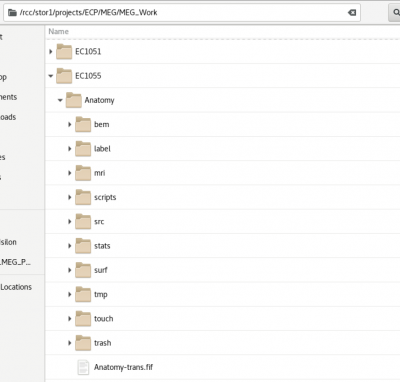
Data Layout
RAW and tSSS
The data is labelled with Subjid+_+DataType+_run+RunNumber+_raw.fif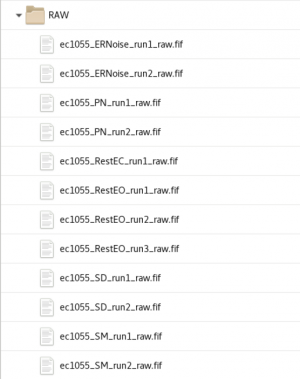
Processing
Initialization - Subject Dataframe
init_subject_dataframe - initialization creates a subject pandas dataframe saved as a csv file in the aProjects folder at the top of the MEG Project folder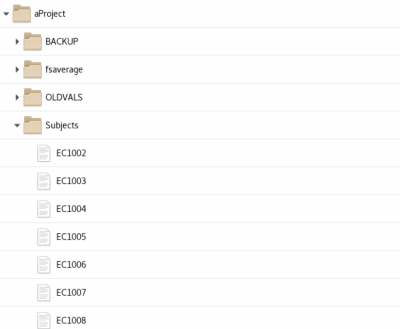
The Contents of the subject dataframe are stored in a .csv file which can be opened using LibreOffice The data file is stored in the row, and all of the documented processing steps are the columns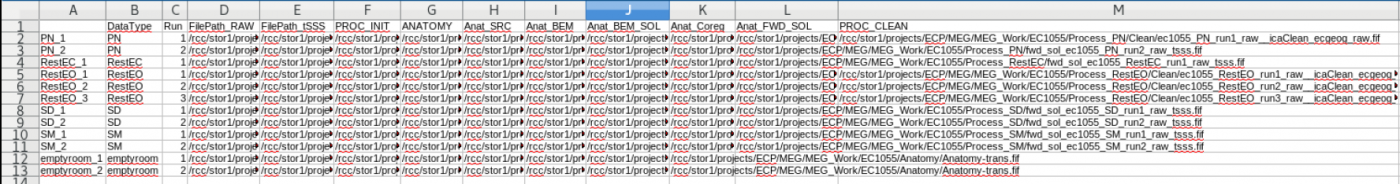
Initialization - Subject Processing Folders
init_subject_process - A process folder is created for each dataset type (multiple runs are copied into each folder) The _tsss.fif files are links to the tSSS folder. If these folders are deleted, it will not affect the data in the tSSS folder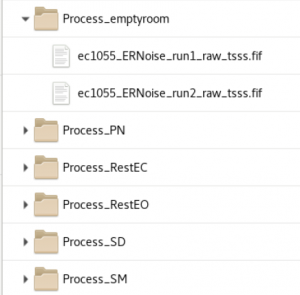
Cleaning
Cleaning (baddata pipeline) performs ICA removal of heartbeat and eyeblink artifact that is correlated to EOG and ECG electrodes at a threshold of (r=???) The processing creates a folder called Process_*DataType*/Clean with the output of the baddata analysis