ECP MEG Data Folders
Jump to navigation
Jump to search
At the MEG lab:
Data is acquired to the MEG_acq/ecp/ folder Data is tSSS processed and organized into raw and tSSS folders (the format of the data can be seen below)Data is uploaded to to the RCC using scp (/rcc/stor1/projects/ECP/MEG/MEG_Orig) Data is copied into /rcc/stor1/projects/ECP/MEG/MEG_Work for further processing
Folder Layout /rcc/stor1/projects/ECP/MEG/MEG_Work
All subjects have a folder with the following folders: Anatomy, RAW, tSSS, Process_*DataType*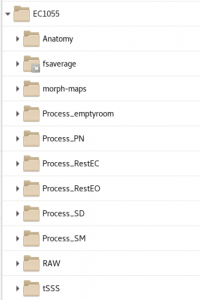
Anatomy Layout
Typical Freesurfer layout with BEM process performed The folder is always labelled Anatomy (a hack to make all folders homogeneous and easier to process) The Anatomy-trans.fif is created during the MNE python coregistration step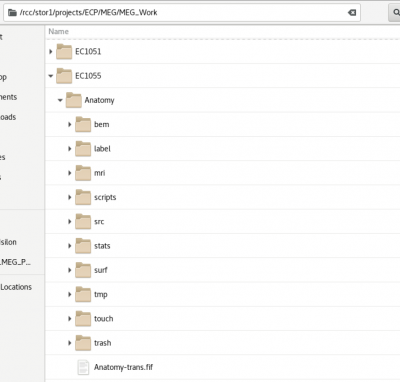
Data Layout
RAW and tSSS
The data is labelled with Subjid+_+DataType+_run+RunNumber+_raw.fif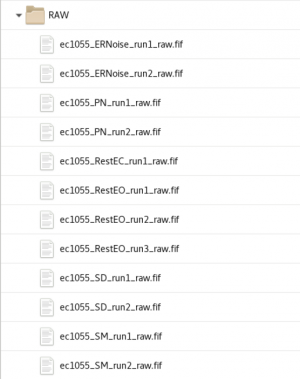
Processing
Initialization
init_subject_dataframe - initialization creates a subject pandas dataframe saved as a csv file in the aProjects folder at the top of the MEG Project folder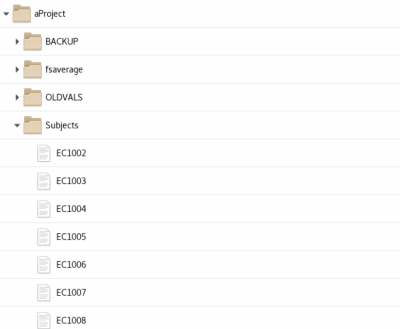
The Contents of the subject dataframe are stored in a .csv file which can be opened using LibreOffice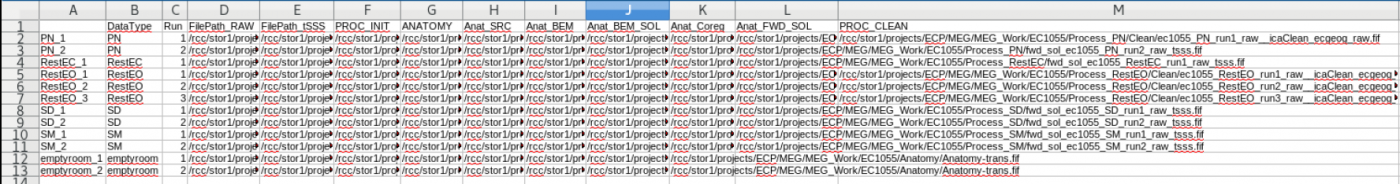
Processing
A set
Cleaning
Cleaning (baddata pipeline) performs ICA removal of heartbeat and eyeblink artifact that is correlated to EOG and ECG electrodes at a threshold of (r=???)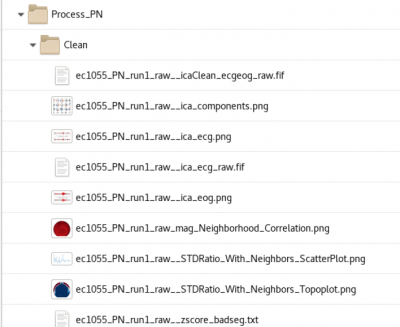
RCC preprocessing
The data is then copied to a folder /rcc/stor1/projects/ECP/MEG/MEG_Work
